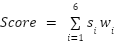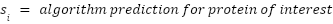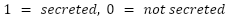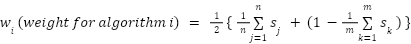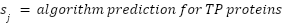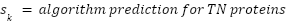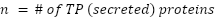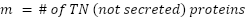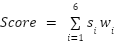
Secretion Prediction Algorithms and Scoring Methodology
True Positive List (verified by literature): 349 secreted proteins
True Negative List (nuclear/cytoplasmic; verified by literature): 612 intracellular proteins
Algorithm | False Negative Rate | False Positive Rate | Description |
0.163 | 0.025 | Consensus prediction (OCTOPUS, Philius, PolyPhobius, SCAMPI and SPOCTOPUS) of membrane protein topology: can separate signal peptide (SP) from transmembrane (TM) region. PMID: 25969446 (2015) | |
0.155 | 0.034 | Hidden Markov Model (HMM) which is a combined TM protein topology and SP predictor. PMID: 15111065 (2004) | |
0.158 | 0.028 | Predicts SP and cleavage position based on a position weight matrix generated from datasets of secreted proteins with experimentally determined cleavage positions. PMID: 15215414 (2004) | |
0.192 | 0.002 | Predicts SP from sequence using dual neural network models, one of which includes proteins with TM regions in the negative training data. 21959131 (2011) | |
0.198 | 0 | Predicts SP and unconventional protein secretion (UPS) in a 2 phase model. A first phase CNN classifier detects SP proteins. A second decision tree classifier trained on in-house experimental data detects UPS proteins. 31857603 (2019) | |
0.229 | 0 | Predicts SP using a CNN and cleavage site with a probabilistic sequence labeling method. 29280997 (2018) |
Score: A custom score is calculated that is the weighted average of the results from all six algorithms. We weight each algorithm by its average prediction accuracy for the input TP and TN lists. If an algorithm produces a probability as output, the authors’ recommended threshold is used for classifying a protein as secreted or not secreted.